Cells in the human body are intricately arranged, forming pathways and spaces for communication, collaboration, and functionality in specific tissues and organs. Any disruption in cell organization can lead to uncontrolled cell growth, cell death, and diseases, such as cancer.
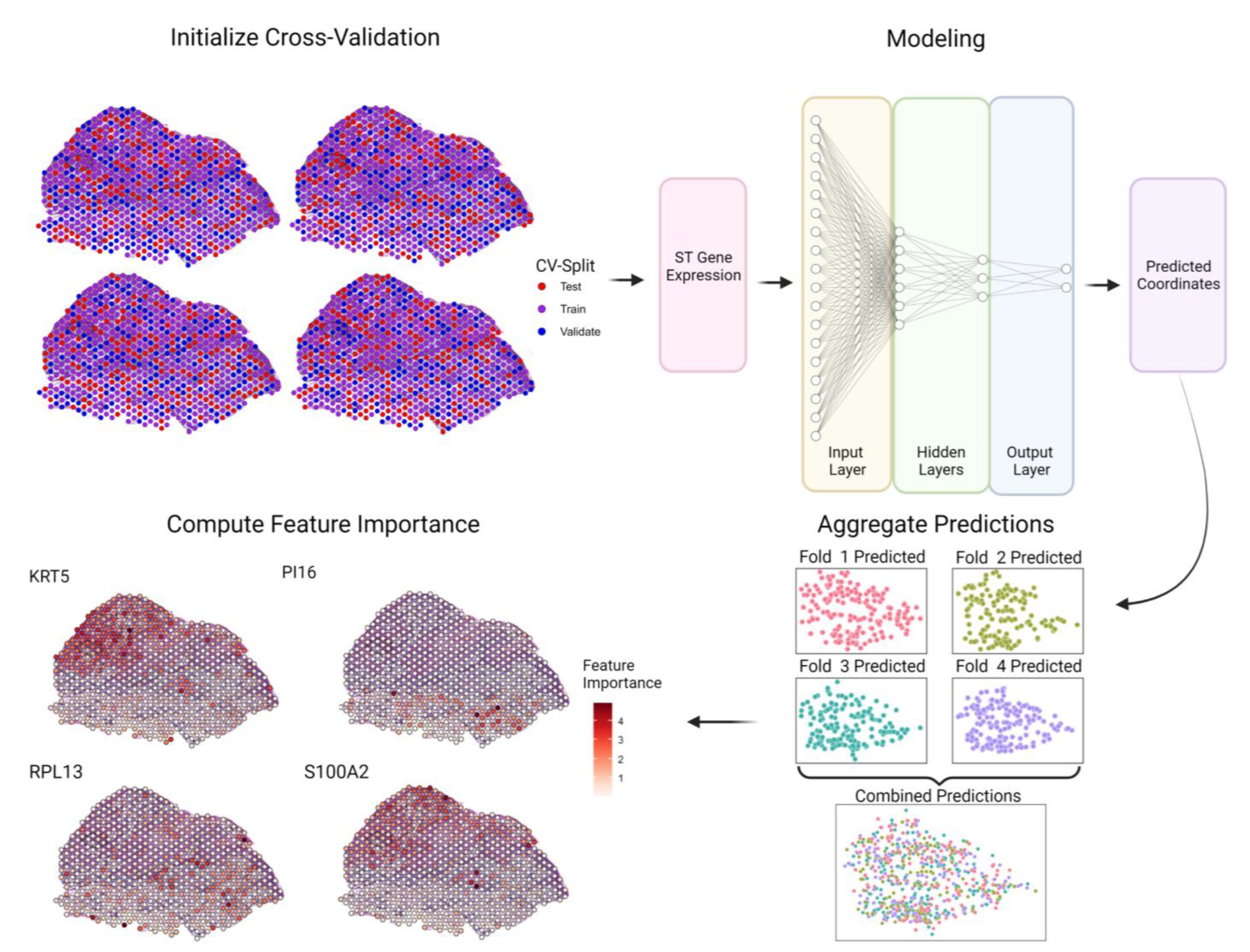
Researchers at the Mayo Clinic Center for Individualized Medicine and Mayo Clinic Comprehensive Cancer Center have developed a cutting-edge artificial intelligence method called Spatially Informed Artificial Intelligence (SPIN-AI). This innovative deep-learning technique can analyze the genetic information of individual cells to reconstruct the precise layout of cells in a tissue, even without prior knowledge of their organization. The study detailing SPIN-AI was recently published in the journal Biomolecules.
To test the effectiveness of their SPIN-AI method, the team used spatial transcriptomic data from squamous cell carcinoma, a type of skin cancer. This specialized data contains information about the active expression of thousands of genes in specific cell locations. By leveraging this data, the researchers generated a detailed map revealing the organization and behavior of cells in the diseased tissue.
“Understanding the organization and communication of cells in diseases is vital for comprehending disease mechanisms and designing new therapeutics,” says Dr. Hu Li, a systems biologist and professor at Mayo Clinic’s Department of Molecular Pharmacology and Experimental Therapeutics and Center for Individualized Medicine. Dr. Li is a co-lead author of the study.
The findings of the study have the potential to pave the way for personalized treatments that target the specific cellular characteristics of each individual.
“We chose this study because the data is readily available and includes high-quality spatial transcriptomic data from numerous patients, which aids in validating our findings,” explains Dr. Cristina Correia, a pharmacology and oncology researcher in Mayo Clinic’s Department of Molecular Pharmacology and Experimental Therapeutics and a co-lead author of the study. “With these findings, we can further investigate ways to manipulate the cellular microenvironment to enhance and sustain drug responses.”
The researchers emphasize that their study was driven by the hypothesis that certain genes can predict cell organization.
Through their SPIN-AI innovation, the researchers identified a new category of genes known as “spatially predictive genes.” These genes provide insights into their activity and location within a cellular niche. Examining these genes allows researchers to gain a deeper understanding of how cells are meticulously organized and the roles they play in specific areas of functionality.
“Many people use AI solely for recognition or classification tasks. However, our work demonstrates that AI can be utilized to make new discoveries driven by specific hypotheses,” states Dr. Choong Ung, a researcher in the Department of Molecular Pharmacology and Experimental Therapeutics. Dr. Ung is also a co-lead author of the study. “Asking the right questions and designing a well-structured training-testing procedure are crucial for harnessing the full power of AI, particularly deep learning,” adds Dr. Ung.
The team plans to continue refining SPIN-AI to explore its potential in identifying and distinguishing different cell populations, as well as investigating strategies to match them with therapeutic targets.
More information:
Kevin Meng-Lin et al, SPIN-AI: A Deep Learning Model That Identifies Spatially Predictive Genes, Biomolecules (2023). DOI: 10.3390/biom13060895
Citation:
AI method predicts how cells are organized in disease microenvironments (2023, June 26)
retrieved 26 June 2023
from https://phys.org/news/2023-06-ai-method-cells-disease-microenvironments.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.
Denial of responsibility! SamacharCentrl is an automatic aggregator of Global media. In each content, the hyperlink to the primary source is specified. All trademarks belong to their rightful owners, and all materials to their authors. For any complaint, please reach us at – [email protected]. We will take necessary action within 24 hours.

Shambhu Kumar is a science communicator, making complex scientific topics accessible to all. His articles explore breakthroughs in various scientific disciplines, from space exploration to cutting-edge research.